Modelling cell level energy density
Thanks to those who positively responded! Happy therefore to share my cell model code here:
So far I have made 1 ‘quickstart’ example of how to build a model similar to that nested in these quote tweets, and more to come later. To quickly summarise:
The code is written in Julia, so you would need that. Simplest way to get started is JuliaPro, and install the necessary add-on packages.
Quickstart example with a basic NMC622/Gr cylindrical cell model is here
Cells are defined as composite object types (structs) which contain other objects (e.g. electrodes, current collectors, electrolyte, separator), all of which have many other properties (e.g. dimensions).
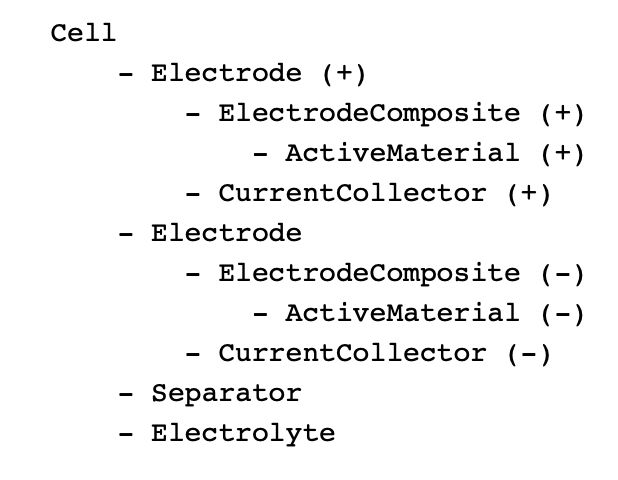
Basic cell model looks like this. Components can be defined as objects, can do maths and specify errors in the definitions of various properties. Cell models can be assigned to objects.

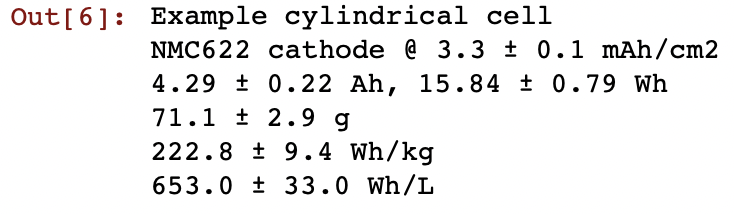
Creating a ‘cell’ object will automatically spit out a simple summary with some basic cell properties. Regular followers will have seen screenshots of these before on various occasions…
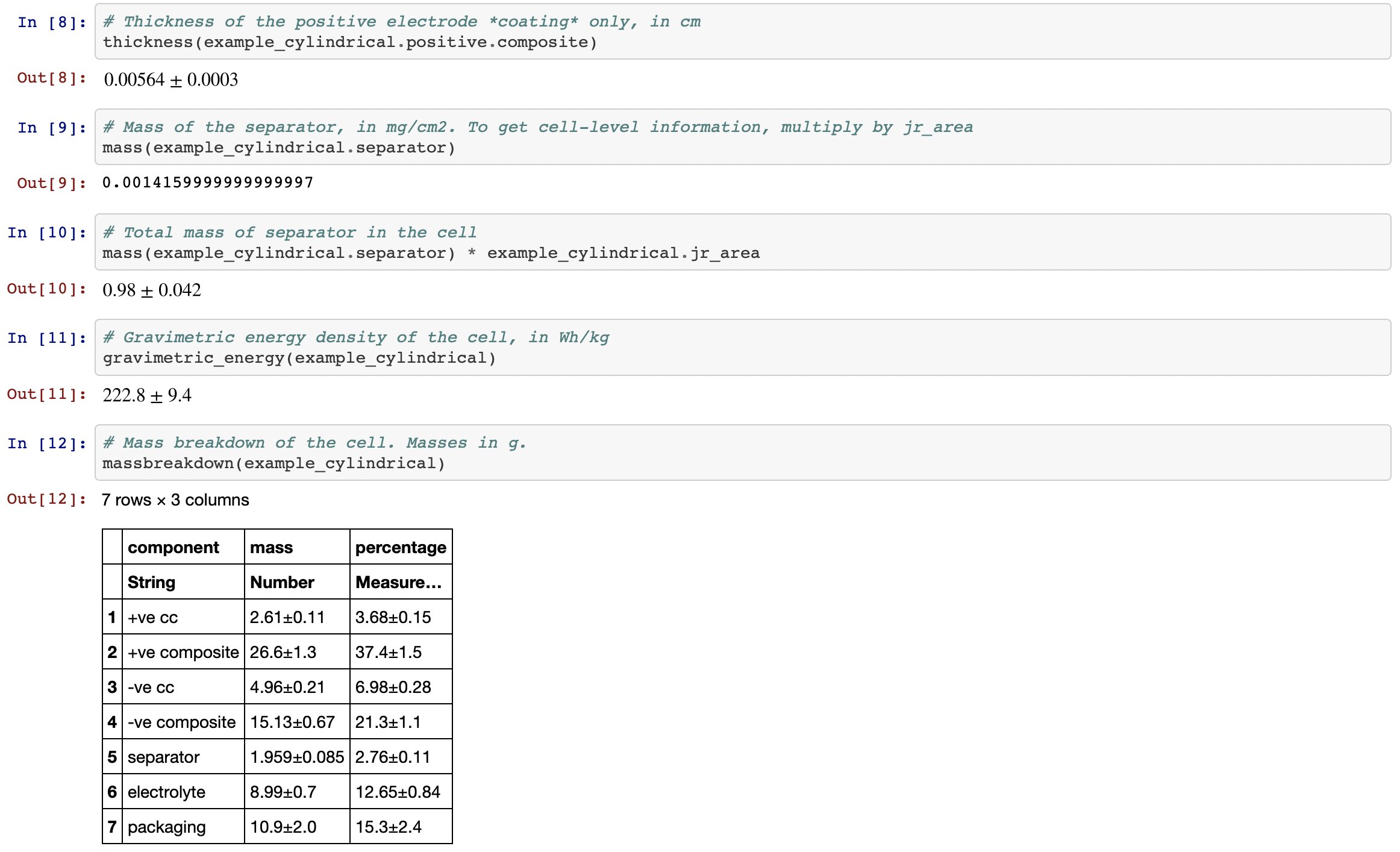
Various other functions exist for extracting numbers/properties of interest. The ‘multiple dispatch’ approach of Julia means that one function (e.g. thickness) can do different things depending on what sort of object you give it (e.g. electrode, or PouchCell)
I will update this when I have a bit more time, to give some more complex examples, and maybe other chemistries, where I have shared numbers before, e.g. solid state. For now, have fun with it!
comments powered by Disqus